Abstract
Tat is an 86 amino acid virally encoded protein vital to the human immunodeficiency virus type 1 (HIV-1) life cycle by stimulating transcription initiation and increases processivity of ribonucleic acid (RNA) polymerase II. Tat is introduced to the endogenous transcription machinery upon binding of the RNA stem-loop TAR. The Tat-TAR interaction is limited to a 6 nucleotide loop and a 3 nucleotide bulge. Given these modest base specific requirements, TAR tertiary structure must serve as the scaffold for Tat binding. A means to study nucleic acid tertiary structure is nuclear magnetic resonance (NMR). Fluorine NMR has a chemical shift range 100-fold larger than the more commonly utilized proton NMR. To generate TAR RNA for 19F-NMR, fluorinated adenosine and uridine nucleotide analogs were incorporated by in vitro transcription in a pUC 19 TAR-Hammerhead expression vector system. Upon transcription with 2F-ATP, 5F-UTP, CTP and GTP, Hammerhead ribozyme autocleavage was inhibited, precluding the production of TAR RNA. Conversely transcription with 2F-ATP, UTP, CTP and GTP resulted in a 31nt TAR transcript, 57nt Hammerhead transcript, and a visible amount of un-cleaved 88nt TAR-Hammerhead transcript, implying decreased ribozyme autocleavage efficiency. Despite decreased autocleavage, milligram quantities of 5F-UTP TAR RNA were synthesized for 19F-NMR ligand binding studies. A model other than the pUC 19 TAR-Hammerhead must be employed to generate 2F-ATP, 5F-UTP TAR RNA.
Introduction
Tat is an 86 amino acid virally encoded protein vital to the human immunodeficiency virus type 1 (HIV-1) life cycle by stimulating transcription initiation and increases processivity of ribonucleic acid (RNA) polymerase II (1,2). Tat is introduced to the endogenous transcription machinery by binding the RNA stem-loop encoded by the trans-activating response element (TAR) (3-5). The TAR element is positioned just distal to the transcription start site conferring a TAR RNA hairpin located at the 5’ ends of mRNA (6). HIV-1 TAR contains two prominent structural features: an apical 6 nucleotide loop and a 3 nucleotide bulge, both requirements for Tat binding (Figure 1) (7). Through extensive mutagenesis it has been shown that Tat binding is localized to the bulge (7) and cellular cofactors bind the loop (8).
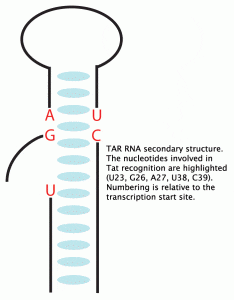 TAR RNA tertiary structure is dynamic. In the absence of ligands, like Tat, an open and accessible major groove is prominent, but a more tightly packed structure is conferred once TAR folds around basic side chains emanating from the Tat protein(9). Given the relatively modest specific-base requirements, (only U23 makes a base-specific interaction with Tat) one infers that TAR tertiary structure must play an active role in protein recognition, and RNA-protein interface formation (10).
TAR RNA tertiary structure is dynamic. In the absence of ligands, like Tat, an open and accessible major groove is prominent, but a more tightly packed structure is conferred once TAR folds around basic side chains emanating from the Tat protein(9). Given the relatively modest specific-base requirements, (only U23 makes a base-specific interaction with Tat) one infers that TAR tertiary structure must play an active role in protein recognition, and RNA-protein interface formation (10).
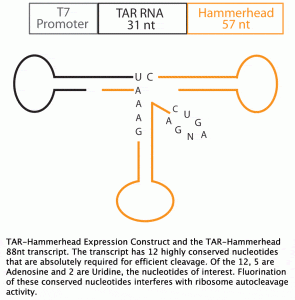
A means to study nucleic acid tertiary structure is Nuclear Magnetic Resonance (NMR) spectroscopy. Fluorine NMR has an extremely wide range of chemical shifts, nearly a 100-fold larger than that of proton NMR, implicating 19F NMR as a valuable tool in ligand binding studies, such as Tat-TAR interactions. The first step in employing Fluorine NMR for ligand binding studies is the incorporation of fluorinated nucleotides. In addition, TAR RNA transcripts with defined 5’ and 3’ ends are required for NMR analysis. To satisfy this requirement, we employed a TAR-Hammerhead construct, where the Hammerhead ribozyme autocleaves itself from the transcription product, resulting in a TAR transcript and a Hammerhead transcript (11). The experimental goal of this study is to incorporate 2F-Adenosine and 5F-Uridine into HIV1-TAR RNA by in vitro transcription using the pUC 19 TAR-Hammerhead plasmid.
Methods and Materials
Template DNA and Fluorinated Nucleotides
Fluorinated Adenosine (2F-Adenosine) nd Uridine (5F-Uridine) nucleotide analogs were obtained from colleagues at the Scripps Research Institute in La Jolla, California (Figure 2). The 2F-Adenosine, 5F-Uridine, Cytidine and Guanosine solution concentration was 114.1 mM. A separate 54.9 mM solutionwas acquired containing independent 5F-Uridine. The pUC 19 test plasmids carried a 31 nucleotide HIV-1 TAR sequence (GGCCAGATTTGAGCCTGGGAGCTCTCTGGTC) and a 57 nucleotide Hammerhead ribozyme sequence (GACGGCTTCGGCCGTCCTGATGAGTCCGTCCTGATGA
GTCCGTGAGGACGAAACCAGAGAGCTCCGGATCC) in the multiple cloning site downstream of the T7 polymerase promoter sequence (TAATACGACTCA CTATA). The plasmid solution was acquired from colleagues at the Scripps Research Institute in La Jolla, California.
 Transcription Condition Optimization
Transcription Condition Optimization
Prior synthesis of milligram quantities of TAR, 24 20ul pilot transcriptions were run in order to assess optimal nucleotide and magnesium conditions for RNA synthesis. The optimization followed a 3 x 4 matrix: 3 nucleotide concentrations varied across 4 magnesium concentrations. The variable nucleotide concentrations were 21mM, 24 mM, and 27 mM. The variable nucleotide concentrations were 21mM, 24mM, 27mM, 30mM.
The optimal conditions for RNA synthesis were qualitatively determined by fractionating the transcription products using 15% polyacrylamide gel electrophoresis.
Unfluorinated in vitro Transcription: Control
The control transcription reaction mixture (15 ml) contained 750 μg template DNA, 21 mM standard nucleotides, 30 mM MgCl2 , 10 mM dithiothreitol (DTT), 75 μl 5000 U RNase Out (invitrogen), 3112.5 μg T7 RNA polymerase, 1X Transcription buffer (400 mM Tris pH 8.0, 100 mM DTT, 10 mM Spermidine FW 145.25, 0.1% Triton X-100) and diluted with 18.2 ohm water. The transcription reaction mixture was incubated for 3 hours at 37 o C. Control RNA transcripts were fractionated by electrophoresis on 15% polyacrylamide gels containing 8 M Urea, .89 M Tris base, .89 M Boric acid, 20 mM EDTA ( pH 8.4). The gels were commassie stained and photographed on a Kodak 4000 MM Image Station.
Fluorinated Flavors of RNA
The 5F-UTP, 2F-ATP, GTP, CTP transcription reaction mixture (20 μl) contained 4.15 μg template DNA, 21 mM fluorinated nucleotide solution, 30 mM MgCl2, 10 mM DTT, .1 μl 5000 U RNase Out (invitrogen), 4.15 μg T7 RNA polymerase, 1 X Transcription buffer and diluted with 18.2 ohm water. The fluorinated transcription reaction mixture was incubated for 3 hours at 37 o C.
The 5F-UTP, ATP, GTP, CTP, transcription reaction mixture (15 ml) contained 750 μg template DNA, 5.25 mM ATP, 5.25 mM CTP, 5.25 mM GTP, 5.25 mM 5F-UTP, 30 mM MgCl2, 10 mM DTT, 75 μl 5000 U RNase out (invitrogen), 3112.5 μg T7 RNA polymerase, 1 X Transcription buffer and diluted with 18.2 ohm water. The fluorinated transcription reaction mixture was incubated for 3 hours at 37 o C. Fluorinated RNA transcripts were fractionated and photographed by the previously described methods.
Results
TAR-Hammerhead Synthesis and Autocleavage
Gel A contains transcripts from the unfluorinated control transcription, with 31 nt TAR and 57 nt Hammerhead separated, implying complete Hammerhead ribozyme autocleavage. Gel B contains transcripts from the fluorinated nucleotide transcriptions, with lane 1 as a size marker and lane 2 as an unfluorinated control. Lane 3 shows transcription products from the 5F-Uridine transcription. Near the bottom is the 31 nucleotide TAR RNA. In the center is the 57 nucleotide hammerhead ribozyme. Interestingly, near the top is the 88 nucleotide Hammerhead + TAR construct, indicating less efficient cleavage of the hammerhead ribozyme than that of the unfluorinated control. Conversely, lane 4 shows the solitary 88 nucleotide TAR-Hammerhead construct, implying no autocleavage activity by the ribozyme with incorporated 2F-Adenosine and 5F-Uridine.
Discussion and Conclusion
TAR is a virally encoded RNA that binds the viral 86 amino acid Tat protein. Once bound, the RNA-protein interface associates with the endogenous transcription machinery, implicating the Tat-TAR interaction as an integral part of the HIV life cycle. Indeed Frankel et al. have shown that the Tat-TAR interaction and subsequent transcription machinery association is absolutely required for efficient transcription and, ultimately, viral replication. Given the integral role of the Tat-TAR interface in the HIV life cycle, understanding the physical dimensions of the interaction could potentially provide insights for anti-HIV therapy.
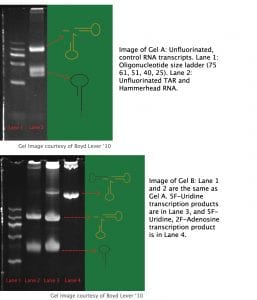 The incorporation of fluorinated nucleotide analogs into TAR RNA is the first step toward utilizing fluorine NMR in ligand binding studies. Incorporating fluorinated Adenosine and Uridine into TAR allows for the positioning of fluorine probes in the major and minor grooves of the TAR structure. Given that RNA polymerases in general, and the T7 RNA polymerase utilized in this study have non-templated transferase activity, a transcription model that generates TAR RNA with defined 3’ and 5’ ends is necessary. Given the Hammerhead ribozyme’s autocleavage activity and production of defined ends, a construct exploiting this ribozyme is ideal for NMR studies. However, when transcribing TAR with 2F-Adenosine and 5F-Uridine with the pUC 19 TAR-Hammerhead system, no autocleavage by the Hammerhead ribozyme occurs, precluding the production of TAR RNA. Such a transcription renders only the 88 nt TAR-Hammerhead uncleaved transcript. Conversely, transcription with standard GTP, CTP, ATP and 5F-Uridine results in the production of 31 nt TAR transcript, 57 nt Hammerhead transcript, and a visible amount of uncleaved TAR-Hammerhead 88 nt transcript.
The incorporation of fluorinated nucleotide analogs into TAR RNA is the first step toward utilizing fluorine NMR in ligand binding studies. Incorporating fluorinated Adenosine and Uridine into TAR allows for the positioning of fluorine probes in the major and minor grooves of the TAR structure. Given that RNA polymerases in general, and the T7 RNA polymerase utilized in this study have non-templated transferase activity, a transcription model that generates TAR RNA with defined 3’ and 5’ ends is necessary. Given the Hammerhead ribozyme’s autocleavage activity and production of defined ends, a construct exploiting this ribozyme is ideal for NMR studies. However, when transcribing TAR with 2F-Adenosine and 5F-Uridine with the pUC 19 TAR-Hammerhead system, no autocleavage by the Hammerhead ribozyme occurs, precluding the production of TAR RNA. Such a transcription renders only the 88 nt TAR-Hammerhead uncleaved transcript. Conversely, transcription with standard GTP, CTP, ATP and 5F-Uridine results in the production of 31 nt TAR transcript, 57 nt Hammerhead transcript, and a visible amount of uncleaved TAR-Hammerhead 88 nt transcript.
The variable cleavage activity of the Hammerhead ribozyme reflects the interference of fluorine with the highly conserved Hammerhead cleavage site. Of the 12 conserved nucleotides 5 are adenosine, thereby increasing the possible interference of fluorine with Hammerhead autocleavage when 2F-Adenosine is present as compared to fluorinated Uridine, which constitutes only 2 conserved nucleotides. Given the relative abundancies of each nucleotide in the conserved cleavage site, one logically assumes that concurrent 2F-Adenosine and 5F-Uridine nucleotides transcriptions would show greater cleavage interference than the use of 5F-Uridine independent of the fluorinated Adenosine. Indeed, we have shown that (i) 5F-Uridine incorporation decreases Hammerhead autocleavage efficiency and (ii) incorporation of 2F-Adenosine and 5F-Uridine inhibits Hammerhead cleavage completely.
Without Hammerhead autocleavage, no TAR RNA is produced when using this model. Thus the cleavage inhibition precludes the concurrent use of the TAR-Hammerhead construct with the 2F-Adenosine, 5F-Uridine analog combination in TAR RNA studies. A different model must be employed to synthesize 2F-Adenosine, 5F-Uridine TAR RNA for ligand binding studies. However, despite decreased autocleavage efficiency, we successfully synthesized milligrams quantities of 5F-Uridine TAR RNA for NMR imaging and future ligand binding studies (data not shown).
Acknowledgements
I wish to thank Mirko Hennig for his generous hospitality in allowing me total access to his laboratory, and the Medical University of South Carolina’s Summer Undergraduate Research Program for their support and instruction.
References:
1.) Feinberg, M.B. , Baltimore, D. and Frankel, A.D. (1991) Proc. Natl. Acad. Sci. USA 88, 4045-4049
2.) Kao, S., Calman, A.F., Luciw, P.A. and Peterlin, B.M. (1987) Nature 330, 489-493.
3.) Keen, N.J., Gait, M.J., and Karn, J. (1995) Proc. Natl. Acad. Sci. USA 93, 2505-2510
4.) Dingwall, C. Ernberg I., Gait, M.J., Green, S.M., Heaphy, S., Karn, J., Lowe, A.D., Singh, M., Skinner, M.A. and Valerio, R. (1989) Proc. Natl. Acad. Sci. USA 86, 6925-6929.
5.) Dingwall, C. Ernberg I., Gait, M.J., Green, S.M., Heaphy, S., Karn, J., Lowe, A.D., Singh, M. and Skinner, M.A. (1990) EMBO J. 9, 4145-4153.
6.) Frankel, A.D. (1992) Curr. Opinion Genet. Dev. 2, 293-298.
7.) Cordingley, M.G., LeFimina, R.L., Callahan, P.L., Condra, J.H., Sandana, V.V., Graham, D.J., Nguyen, T.M., LeGrow, K., Gotlib, L., Schlaback, A.J. and Colonno, R.J. (1990) Proc. Natl. Acad. Sci. USA 87, 8985-8989.
8.) Marciniak, R.A., Garcia-Blanco, M. and Sharp, P.A. (1990) Proc. Natl. Acad. Sci. USA 87, 3624-3628.
9.) Fareed Aboul-ela, Karn, J. and Varani, G. (1996) Nucleic Acids Research 24, 3974-3981.
10.) Frankel, A.D. (1992) Protein Science 1, 1539-1542.
11.) Pley, H. W., Flaherty, K. M. and McKay, D. B. (1994) Nature 372: 68-74.