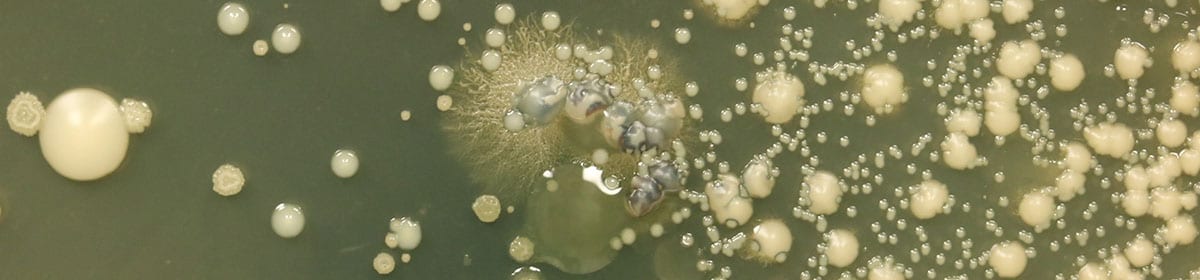
Our goal is to understand how microbial cells interact with one another in the context of human disease. Research areas include bacterial-fungal interactions, interactions within genetically heterogeneous microbial populations, and the development of new ways to analyze complex microbial communities and populations in clinical samples. Below are examples of our on-going projects.
Candida fitness in chronic infections
Candida, yeasts that can live benignly in host-associated microbial communities, can also cause serious localized and disseminated infections. We are interested in how Candida evolution can reveal important information about how yeast persist and grow in the host. We are specifically focused on how studying the natural heterogeneity within and between chronic infections can help us identify new ways to prevent, mitigate or eliminate infections. Our current work on how Candida lustitaniae, a haploid Candida species not frequently found in the human microbiota, evolves as it adapts to the host environment. C. lusitaniae is phylogenetically closely related to Candida auris, a recently recognized pathogen that has repeatedly developed drug resistance and causes outbreaks in health care facilities. We hope to use the study of these two species to learn about how fungal pathogens emerge.
Recent papers focus on on heterogeneity and tradeoffs associated with the fungal evolution of drug resistance, and other roles for drug resistance regulators in diverse Candida species including Candida auris.
Pseudomonas aeruginosa fitness in chronic infections
Pseudomonas aeruginosa causes a wide range of infections and is a particularly problematic pathogen of the lung in compromised individuals and in individuals with the genetic disease cystic fibrosis (CF). We are interested in understanding how as of yet uncharacterized pathways increase fitness in the microoxic and metal-restricted environments found in infections, and how naturally-occurring P. aeruginosa variants have improved fitness in these environments. We are particularly interested in how the host environment and within-host evolution shapes both inter- and intra-species interactions. Our recent studies have focused on how LasR- variants of P. aeruginosa interact with their LasR+ ancestors leading to emergent properties for genetically heterogeneous populations.
Machine learning analysis of microbial gene expression
 We are collaborating with Casey Greene in the development a machine learning-derived model for the analysis of P. aeruginosa gene expression using publicly available gene expression data. Our goals include learning more about how P. aeruginosa and other bacteria change in the presence of other microbes and in the host without relying on previously determined reference conditions, and to make the large amounts of publicly available data more accessible to scientists regardless of their computational skill sets. Click here to access our eADAGE model for P. aeruginosa gene expression.
We are collaborating with Casey Greene in the development a machine learning-derived model for the analysis of P. aeruginosa gene expression using publicly available gene expression data. Our goals include learning more about how P. aeruginosa and other bacteria change in the presence of other microbes and in the host without relying on previously determined reference conditions, and to make the large amounts of publicly available data more accessible to scientists regardless of their computational skill sets. Click here to access our eADAGE model for P. aeruginosa gene expression.
Characterization of populations and gene expression in the CF lung
Our recent work has developed methods and tools for the characterization of bacteria and fungi in the CF lung and other clinical samples. We have developed strategies for the analysis of microbial gene expression using the NanoString technology in clinical samples, an improved database for the analysis of fungal populations, an RNA based method for profiling microbial communities. We have recently collaborated with the lab of Carey Nadell to study Pseudomonas-Candida dynamics in a microfluidics model which informs approaches to analyze complex communities in clinical samples.