Many processes in a living cell are regulated through the specific interaction between proteins. Targeting the disruption of protein-protein complexes for therapeutic purposes is considered a very attractive, albeit difficult, approach. In our research, we take advantage of information on the protein’s overall fold, primary and secondary structure at the interface and flexibility to identify putative binding sites and design and test molecules that interfere with or block protein-protein interaction. We focus on peptides, peptidomimetics, and small molecular weight compounds.
Nuclear magnetic resonance (NMR) provides the unique advantage of allowing to investigate both protein structure and dynamics and has been effectively employed as a screening technique, especially in fragment-based approaches. We complement our NMR studies with a number of biophysical and biochemical techniques for characterization of proteins and binding interactions (EPR, CD, fluorescence, etc.) and with computational tools for structural refinement and model building.
The NEMO project 
This NIGMS R01 funded project is the closest to my heart and a major thrust of our research.
The NF-κB essential modulator (NEMO) is an essential component in the activation of the canonical NF-κB pathway and exerts its function by recruiting the inhibitor of κB kinases IKKα and IKKβ to the IKK complex. Inhibition of the interaction between the IKKα and IKKβ kinases and NEMO is considered an attractive therapeutic avenue to the inhibition of NF-κB, due to the pathway’s role in human diseases, encompassing inflammatory and autoimmune diseases and cancer.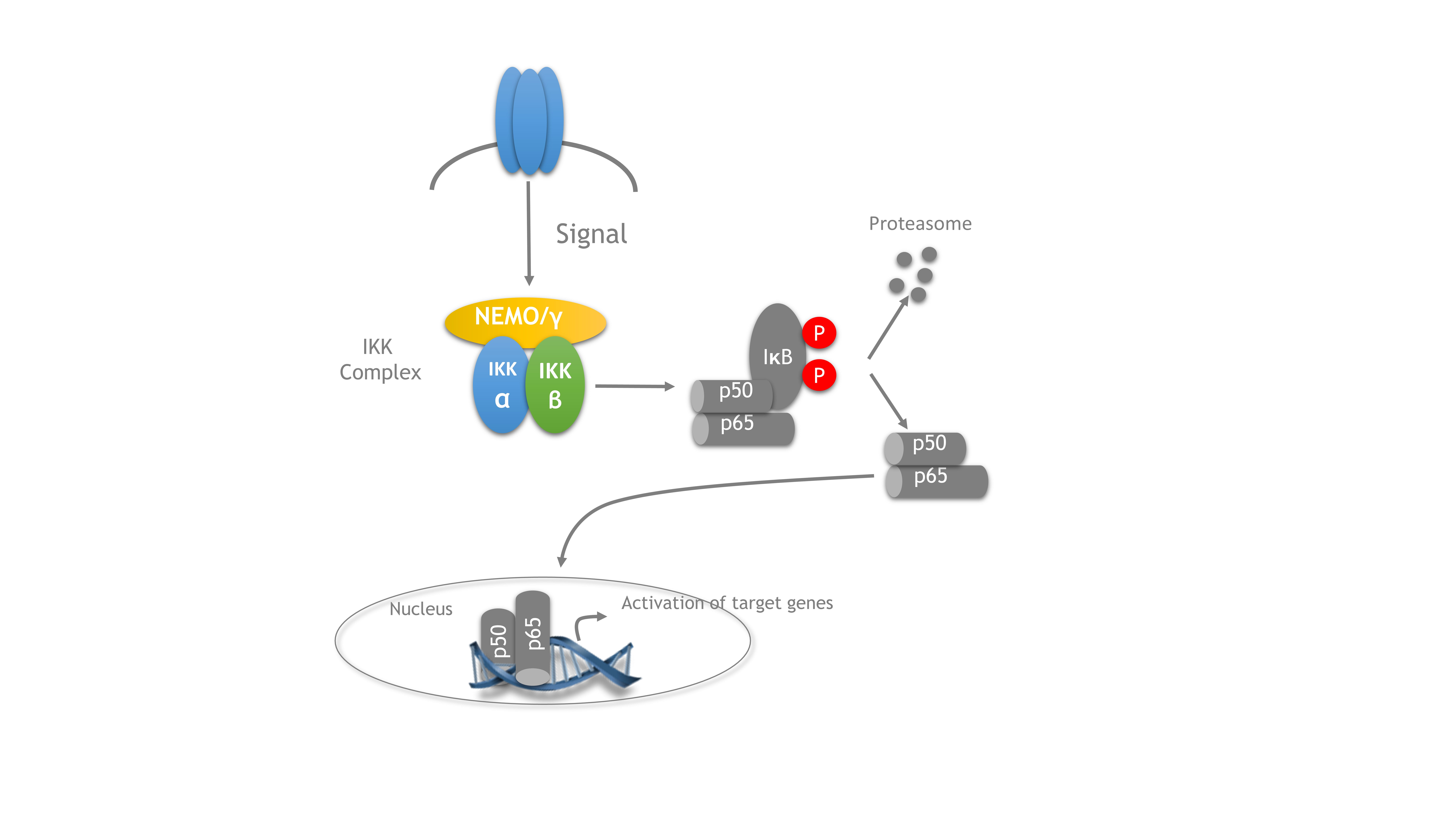
The overall goal of the project is the understanding of the mechanism for NEMO inhibition and its exploitation for the development of small molecule inhibitors. The long-term expectation of the project is to establish the structural basis for inhibitor design and the requirements for high affinity binding and specificity. We pursue this through different approaches. We are actively working on the determination of the structure of NEMO-inhibitor complexes, which rely on a new construct of the IKK-binding domain of NEMO which easily yields X-ray structures. 
We have published our results on the engineering of the IKK-binding domain of NEMO, which allowed us to determine the structure of unliganded NEMO(44-111) by X-ray crystallography.
Guo B, Audu CO, Cochran JC, Mierke DF, Pellegrini M. Protein engineering of the N-terminus of NEMO: structure stabilization and rescue of IKKβ binding. Biochemistry. 2014 Nov 4;53(43):6776-85. doi: 10.1021/bi500861x. Epub 2014 Oct 23. PMID: 25286246; PMCID: PMC4222529.
Barczewski AH, Ragusa MJ, Mierke DF, Pellegrini M. The IKK-binding domain of NEMO is an irregular coiled coil with a dynamic binding interface. Sci Rep. 2019 Feb 27;9(1):2950. doi: 10.1038/s41598-019-39588-2. PMID: 30814588; PMCID: PMC6393490.
We have also filmed a really cool how-to video on the production and crystallization of these NEMO constructs:
https://www.jove.com/embed/player?id=60339&access=rqkkvrcz&t=1&s=1&fpv=1
We are working on the rational design of peptide and small molecule inhibitors of the NEMO / IKK interaction . Our lab moves from molecular biology and protein engineering, through protein chemistry and assay development to NMR spectroscopy, X-ray crystallography and computational methods. We screen mostly using in silico methods coupled with NMR-based screening and validate the inhibitors by biochemical, biophysical and cellular methods. We are fortunate to collaborate with @RagusaLab and @HaydenLab!
