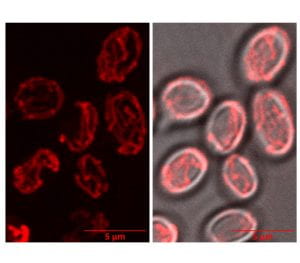
Investigating Histone Control of Cell Size in the Drosophila Embryo
PI: Amanda Amodeo, PhD
Assistant Professor of Biological Sciences
The egg is by far the largest cell type produced by most species. It is so large that the first order of most newly fertilized embryos is to undertake a series of rapid reductive cleavage divisions to reduce the cells to more typical sizes. In many organisms, including our favorite model species, Drosophila, these reductive divisions continue until cells reach the correct ratio of DNA to cytoplasm. Since the genome size remains constant throughout development, but cytoplasmic volume decreases as the cells become smaller the cells can use the genome as a molecular “yardstick” to measure their size. Recently we have uncovered a mechanism by which the embryo uses consumption of a pool of maternally provided histones by the increasing number of embryonic genomes to control cell cycle progression and the cessation of division.
Impact of Pre-existing T Cell Memory on Oncolytic Virus Therapy
PI: Pamela Rosato, PhD
Assistant Professor of Microbiology and Immunology
Oncolytic viruses (OV) are a promising class of cancer therapeutics that work by preferentially infecting and killing tumor cells. Many OVs are viruses to which individuals have pre-existing T cell immunity, through vaccination or natural infection (e.g. HSV-1, measles, and vaccinia virus), yet the impact of this immunity on therapeutic efficacy and patient outcome is unclear. Recent findings have revealed that virus-specific memory T cells populate tumors, often to high frequency. Because of their location within the tumor, it is likely these memory T cells will encounter viral antigen during OV therapy. In light of this, we are interested in understanding the impact of oncolytic virus-specific T cells on OV therapy to then be able to refine and redesign oncolytic viruses for improved efficacy.

Identifying Mitophagy Receptors as Targets in Ras-dysregulated Cells
PI: Michael Ragusa, PhD
(Current) Associate Professor of Chemistry
Pancreatic ductal adenocarcinoma (PDAC) is one of the most lethal and challenging cancers to treat in the United States. The growth of PDAC tumors is dependent on a cellular degradation process, termed autophagy. We are combining biochemical and cellular approaches to uncover the role of autophagy in PDAC.
Saccharomyces cerevisiae were stained with MitoTracker Red CMXRos and imaged using a Zeiss LSM 880 with Airyscan to monitor mitochondrial dynamics.
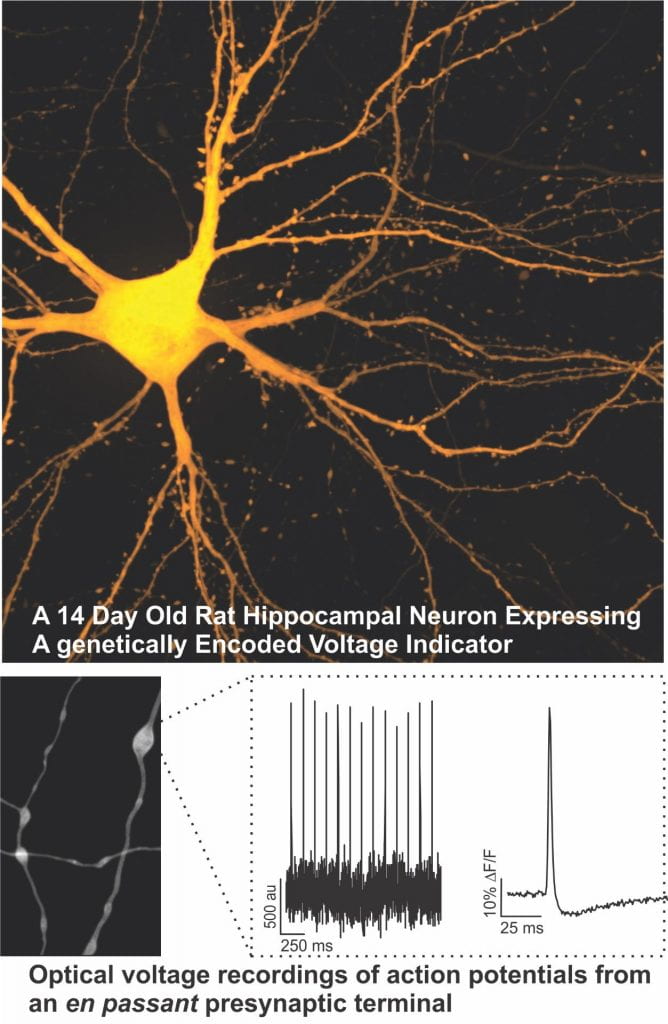
Electrogenic Modulation of Signal Decoding in Presynaptic Terminals
PI: Michael Hoppa, DPhil
(Current) Associate Professor of Biological Sciences
Extramural Funding Awarded: NIH/R01/NSF CAREER
The generation and transmission of electrical signals is fundamental to initiating the release of neurotransmitters in the nervous system. This proposal will take advantage of a number of novel optical approaches to determine the molecular mechanisms that regulate electrical signalling in nerve terminals and define their impact on neurotransmission. Compromised neurotransmission is a known or suspected defect in several neurological diseases, thus a better understanding electrical signaling in nerve terminals could suggest new therapeutic approaches.
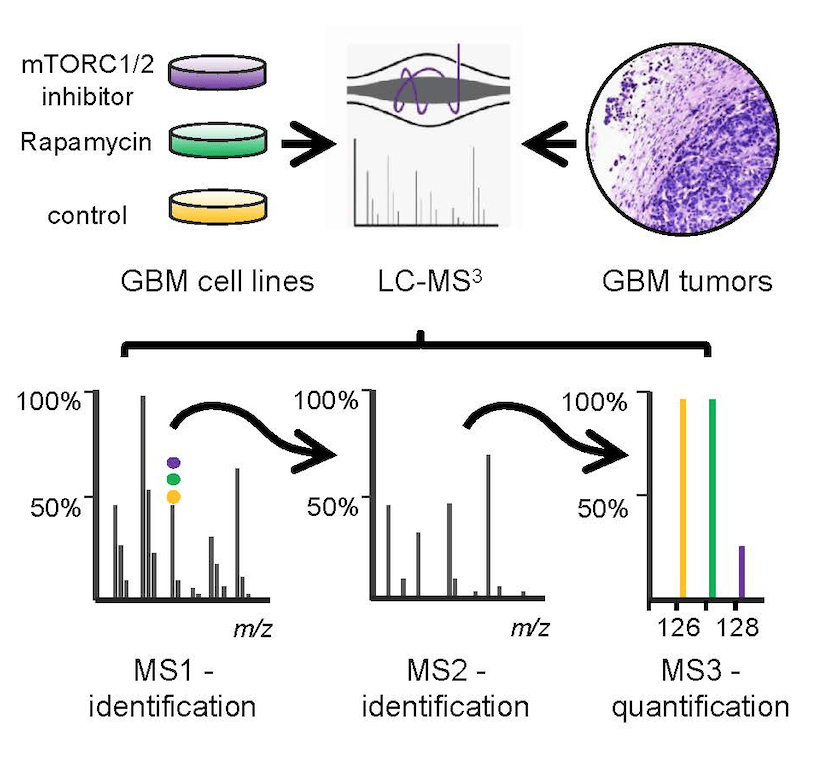
Proteomic Approaches to Target the PP6-mTORC2 Pathway in Glioblastoma
PI: Arminja Kettenbach, PhD
(Current) Professor of Biochemistry and Cell Biology
Extramural Funding Awarded: NIH/R33, NIH/R35
Glioblastoma multiforme (GBM) is the most common and aggressive tumor of the central nervous system in adults and it is generally fatal within 12 months of diagnosis. We propose to identify targets in the newly uncovered and frequently overexpressed PP6-mTORC2 pathway for the development of new therapeutic strategies to fight this devastating disease.
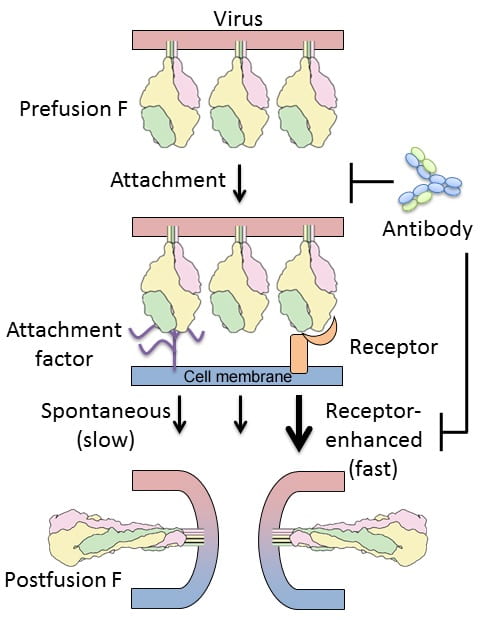
Molecular Mechanisms of RSV F Activation and Inhibition
PI: Jason McLellan, PhD
(Current) Professor, Molecular Biosciences
The University of Texas at Austin
Extramural Funding Awarded: NIH/R01, Janssen Vaccines & Prevention B.V., MedImmune
Respiratory syncytial virus (RSV) causes respiratory tract infections that result in substantial morbidity and mortality in infants and the elderly. RSV entry into host cells is facilitated by an attachment protein (G) and a fusion protein (F) that in its active form adopts a metastable prefusion conformation. After attachment of RSV to cells, it is hypothesized that host-factors trigger the conformational rearrangement of F that results in fusion of the viral and cellular membranes. The prefusion conformation of F is therefore considered an ideal vaccine antigen, and antibodies and small molecules that disrupt its structure and function are actively being pursued. Development of effective therapeutics will be greatly enhanced by a molecular understanding of how the F glycoprotein interacts with host-cell factors to promote entry, and how neutralizing antibodies inhibit one or more steps in the entry process. For this project, we will test two hypotheses: first, that specific host-cell factors enhance the rate of RSV F triggering; and second, that the most potent neutralizing antibodies target receptor-binding sites and block conformational changes. We expect that the results from these studies will fill important gaps in our understanding of RSV entry and provide a molecular basis for RSV F activation and inhibition.
Protein Design for Selective Interference with LPA Signaling in Colon Cancer
PI: Gevorg Grigoryan, Ph.D.
Associate Professor of Computer Science
Extramural Funding Awarded: NSF, NIH/R01

Colorectal cancer was the third-most common cause of deaths from cancer in the United States in 2013. This disease claims over 50,000 lives and afflicts over 140,000 people annually in the US, underscoring a clear and urgent need for better control of this incurable malignancy. The goal of our study is to attenuate oncogenic activities in colon cancer cells by inhibiting a key proliferatory signaling pathway mediated by the powerful mitogen lysophosphatidic acid (LPA). Key towards this goal is to produce selective inhibitors—molecules that interact with and inhibit specific proteins in the cell, while avoiding any unintended side-interactions with other proteins. We are uniquely capable of providing such selectivity using computational technologies we have developed that enable us to target selectively a single member in a family of closely related protein domains.
Mechanisms of Increased Lung Disease Secondary to Polymorphisms in the ACE Gene
PI: Alix Ashare, M.D., Ph.D.
Associate Professor of Medicine and Microbiology & Immunology
Extramural Funding Awarded: NIH/R01, Cystic Fibrosis Foundation, American Asthma Foundation
Current treatments for chronic obstructive pulmonary disease (COPD) attempt to ameliorate symptoms and prevent exacerbations. Studies have identified an insertion (I)/deletion (D) polymorphism in an intron of the angiotensin converting enzyme (ACE) gene. D/D homozygotes (~25% of the population) exhibit more severe COPD but the mechanism is unclear. Our major goal is to investigate and target the role of ACE in the development of COPD. Patients with COPD are plagued by chronic inflammation, leading to increased lung compliance. Alveolar macrophages (AMs) are critical to the local inflammatory response in COPD. ACE is expressed on the surface of AMs and its abundance regulates levels of angiotensin II (ANGII), which in turn binds the ANGII type I (AT1) receptor on AMs. Inflammation in COPD causes bronchospasm, which can result in hypoxia. Hypoxia, in turn, causes an increase in ANGII, a key inflammatory mediator, creating a vicious cycle. Our preliminary data show that AMs generate an inflammatory response to hypoxia that is blocked by the AT1 receptor blocker, losartan. We also found increased pulmonary ACE and ANGII in subjects with the ACE D/D genotype. Our data suggest that the intronic sequence encodes microRNAs (miRNAs), small RNAs that can regulate protein abundance. Our overarching hypothesis is that the ACE D/D genotype increases translation of ACE protein, leading to an exaggerated ANGII response during hypoxia, increasing the severity of COPD. Aim 1 will test the hypothesis that the ACE D/D polymorphism is associated with elevated ACE activity and lung inflammation compared to I/I subjects. In parallel, we will determine if these levels predict differences in lung compliance, as quantified by magnetic resonance elastography (MRE). In Aim 2, we will test the hypothesis that hypoxia causes a dose-dependent increase in ACE and ANGII that is exaggerated in AMs from D/D versus I/I subjects. Our preliminary data suggest the expression of ACE-specific miRNAs from the intronic sequence. Aim 3 will test the hypothesis that reduced expression of these miRNAs in D/D subjects causes an increase in ACE translation compared to I/I subjects. This will foster discovery of previously unrecognized targets that could decrease ACE activity in the lung.