The goal of this project was to develop network-based methods to predict tissue-specific pathways that underlie diseases and drug responses. Successfully treating systemic diseases requires targeting diverse, tissue- specific disease processes, which are not easy to measure directly. Internal organ biopsies are rare and almost never taken in healthy subjects due to their inherent risks. Experiments probing disease states and drug responses with high-throughput (HTP) gene expression have to mediate a tradeoff between the accessibility and tractability of the assayed biological system and the direct translatability of results to target tissues. For example, peripheral tissues such as skin and blood are easily acquired and typically contain important information about the pathobiology of diseases, but HTP data in peripheral tissue is not a perfect surrogate for HTP data in other tissues. Likewise, drug screening in cell culture allows for rapid and scalable determination of gene-expression-response signatures to therapeutic compounds, but translating these results to target tissues is not straightforward.
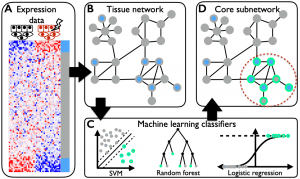
The overarching goal of this project was to predict tissue-specific pathways from easily obtained HTP data from outside that tissue. This project sought to develop and validate a novel machine- learning framework called “tissue network knowledge transfer” (TINKER), which predicts tissue-specific pathways using HTP data from outside that tissue by mining tissue-specific gene-gene interaction networks. TINKER was used to predict differential gene expression in internal organs from HTP gene signatures obtained from skin and blood from the same disease condition. TINKER was tested by using it to predict known drug targets in tissues from HTP gene signatures in cell culture. Finally, this project aimed to optimize TINKER by incorporating nonlinear machine learning algorithms and network feature representations that incorporate indirect connections among genes.