Imagine that you receive a reminder from the doctor’s office for an annual checkup. After mulling it over, you make an appointment to see your primary care provider. She runs through a routine physical and notices nothing amiss, but based on your family history of cancer as written on your digital health record, she pauses to feel for abnormal growth.
Sure enough, your physician detects a lump on your neck, a lump so small that you hadn’t noticed it before. After administering a local anesthetic, she extracts a small amount of fluid from the suspicious area and sends the specimen off to the laboratory for a biopsy.
A day later, your physician delivers the bad news: you have cancer. Fortunately, based on a genetic analysis of your specimen, the laboratory has determined the exact subtype of cancer and the known mechanisms that spur on its growth; the prognosis is positive. Moreover, using a genetic database containing millions of known genetic reaction to chemical compounds, your physician has compiled a list of possible treatments that maximizes efficacy with minimum harmful side effects.
And most, if not all, of the costs of diagnosis and treatment is covered by your insurance provider.
As of now, the above scenario seems suited for a futuristic medical utopia, when various components of health care—from diagnosis to reimbursement—will be tailored to the individual patient. But developments are already underway towards personalized medicine: a medical model that allows for the customization of healthcare, with all decisions and practices tailored to the individual patient via use of genetic information.
Currently, personalized medicine exists only in fragments, at varying stages of development. For the full realization of personalized medicine for all, scientific development, medical translation, health delivery and health policy must be coalesced into a streamlined form.
Personalized Medical Diagnosis
The Human Genome Project
In 2003, the Human Genome Project completed its primary goal of mapping the approximately 20,000 to 25,000 genes of the human genome. The project’s completion led to a flurry of speculation of the future of medicine incorporating genomic analysis. While we have yet to meet these expectations, advances in medical genetics have enabled a more detailed understanding of the impact of genetics in disease.
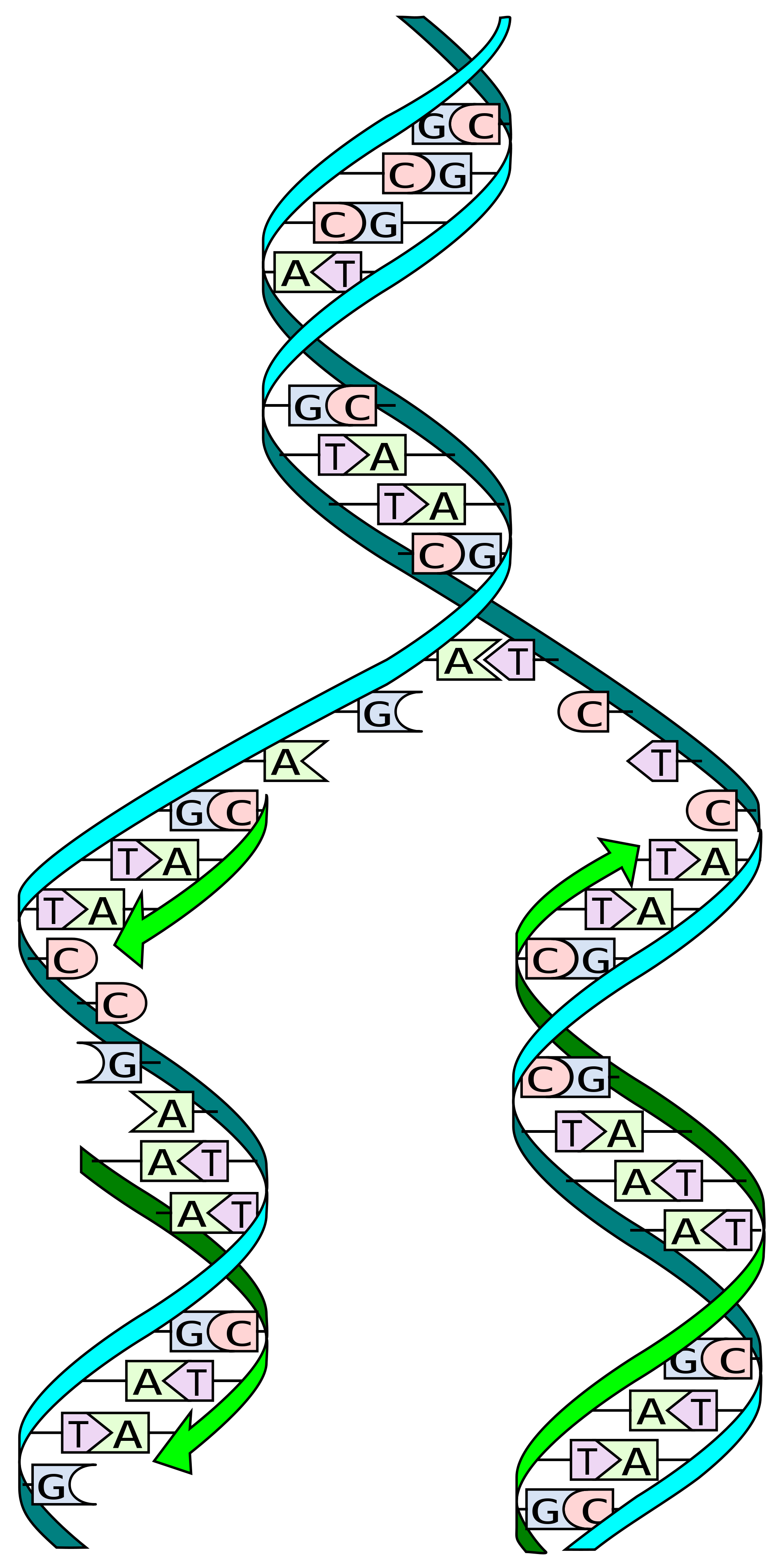
Figure 1: Schematic of DNA replication, a process investigated for the Human Genome Project.
Specifically, the Human Genome Project revealed the importance of single nucleotide polymorphisms (SNPs) in accounting for the genetic variability among individuals (Fig. 1). More specifically, Genome-Wide Association Studies (GWAS) have enabled the examination of genetic variations and their effects; by analyzing common genetic variants in different individuals, variants associated with complex traits—particularly diseases—can be identified (1).
However, GWAS is not without criticism. Opponents have criticized GWAS of insufficiently explaining genetic variation in populations and for not delivering much knowledge of clinical utility. Proponents have pointed out the GWAS design in human populations “has led to new discoveries about genes and pathways involved in common diseases and other complex traits, provided a wealth of new biological insights, led to discoveries with direct clinical utility, and facilitated basic research in human genetics and genomics.” Furthermore, future technological advances will allow entire genomes to be sequenced for affordable prices, generate additional genes, pathways, and biological insights, and identify causal mutations (1).
Thus, despite its criticisms, the Human Genome Project and GWAS have laid the scientific foundations and set off the public imagination on the possibility of personalized medicine.
Whole Genome Sequencing
Last July, the New York Times published a story about cancer biologist Dr. Lukas Wartman, a genetic researcher at the Washington University in St. Louis who had developed Acute Lymphoblastic Leukemia (ALL), the very cancer that he had been studying in his lab.
Fortunately for Dr. Wartman, his institute came to his aid. Dr. Timothy Ley, the associate director of Washington University’s genome institute, called upon his lab to see whether they could find a “rogue gene” that was spurring Dr. Wartman’s cancer. To observe the inner workings of his body, the team sequenced the complete genomic makeup of both his cancerous and healthy cells. By comparing the two sequences, the team deduced that a mutation had caused a gene to be overexpressed; the researchers even identified a promising new drug that might turn off the rogue gene causing the cancer.
While the drug was originally tested and developed for advanced kidney cancer patients, Dr. Wartman used it to successfully treat his leukemia. He is now in remission (2).
While whole genome sequencing is currently available to the public on a small scale, experts believe that its widespread use may not be too far off in the future. Until then, more specific and limited means of genomic diagnosis are available.
Currently available diagnostics
Molecular diagnosis already has a rich precedence in oncology. Novel molecular experimental methods have enabled tests for markers, genes, proteins, and protein pathway activation expression profiles; these, combined with identification of somatic mutations in cancer cells, have allowed for better defined prognosis and suggestions of effective treatment options.
For a diagnosis assessing an individual’s risk to both common and uncommon diseases, a small-scale personalized diagnosis through DNA analysis is currently available. Biotechnological companies have achieved this by synchronizing the rapidly growing body of pharmacogenomic knowledge with commercial gene sequencing and analysis.

Figure 2: 23andMe is a biotechnology company that provides personal genetic testing for consumers.
For instance, 23andMe, a Silicon Valley biotech firm, provides rapid genetic testing and counseling for the concerned and the curious who have the money to spend (Fig. 2). Founded by Anne Wojcicki, wife of Sergey Brin, the company has been profiled by popular publications such as Collins’ “The Language of Life: DNA and the Revolution in Personalized Medicine” and Time, which awarded 23andMe the 2008 “Invention of the Year.”
For a flat fee of $299, the company ships its customers a kit to collect saliva samples. Within 2-3 weeks of the kit’s receipt, the customers receive an analysis of their personal genetic variations over the Internet, including carrier status, disease risk, and drug response. While 23andMe has resisted government regulation for years, the company has moved to ask the Food and Drug Administration to approve its drug test as a medical service, which, if successful, may boost scientific credibility and acceptance within medical and scientific circles that have questioned its usefulness (3, 4).
Yet, while 23andMe offers historic, unprecedented access to the possible implications of one’s genetic information, genes does not necessarily equate to destiny. Scientists have long debated whether disorders are written into our genetics (nature) or picked up through environmental exposure (nurture). Today’s scientific consensus allows for both through the concept of epigenetics, in which environmental factors—ranging from molecular methylation and acute chemical exposure to long-term diet and lifestyle choices—can affect the expression of our genes.
The Development of Personalized Therapies
Population-Based Medicine, A step towards personalized medicine?
Historically, the crossing of science and race evokes centuries of scientific racism in which the vogue scientific methods of the time—ranging from phrenology to physical anthropology to modern-day genetics—were applied to justify racial oppression.
Yet, the scientific community has once again raised the question of race in a way that may prove beneficial to all—race-based medicinal therapies. By stepping away from the current one-size-fits-all modality, race-based medicinal therapies can target specific populations and provide a stepping stone towards personalized medicine. However, these developments may come at a cost: race-based therapies can have unintended side effects on our social fabric.
Tay-Sachs Disease: Community-targeted therapies
Race-based therapies are already utilized within populations facing increased genetic susceptibility to medical conditions. For instance, Tay-Sachs disease is an autosomal recessive neurodegenerative disorder in which harmful quantities of a fatty substance build up in the brain due to insufficient activity of an enzyme that catalyzes the biodegradation of the fatty substance (5). The disorder is observed primarily in Cajun, French Canadian, and Ashkenazi Jewish populations. The first program to prevent Tay-Sachs originated decades ago in Jewish synagogues and community centers of Baltimore and Washington D.C., which pushed for testing of potential carriers. The idea spread to cities throughout the United States and led to the virtual prevention of Tays-Sachs disease within the Jewish population through mate selection (6).
Population-targeted studies are nothing new in epidemiology, considering that members of certain ethnicities share similar diets, values, lifestyle choices, and genetic traits. However, problems arise when science correlates race with genetics.
BiDil: Drugs for African Americans
Race-based therapies have also appeared in the pharmaceutical industry. In June 2005, the F.D.A. approved the first drug to be intended for one racial group—African Americans. BiDil, manufactured by Nitromed, is a combination of two generic vasodilator drugs (isosorbide dinitrate/hydralazine hydrochloride) that prevent heart failure by relaxing blood vessels.
The effects of the drug combination were observed under the African American Heart Failure Trial (A-HeFT), which demonstrated a 43 percent reduction in mortality rate for African Americans’ heart failure patients treated with the dual-drug combination (7).
The potent combination of race and medicine has also raised academic backlash. Following the study’s publication in the New England Journal of Science, ethicists pointed out that A-HeFT enrolled only self-identified African Americans, which may be a reflection of socio-cultural—rather than strictly genetic—characteristics. Moreover, scientists contended that because the study did not investigate the effects of the drug combination on a non-African American population, researchers cannot convincingly conclude that BiDil works unilaterally on the African American population (8). Perhaps most concerning to society at large, critics have condemned the study for “scientizing” race (8).
Proponents of BiDil have fought back against the allegations, noting that that race-based therapies treat race as a crude marker for genetic variations that have yet to be discovered.
Overall, race-based medicine presents an ethical dilemma. While our public society has strived to minimize racial differences, racialized medicine may enable certain populations to receive more effective treatments.
Pharmacogenomics: Promising personal medicine
While general personalized medicine may be years away, active research is being undertaken to study how an individual’s genetic inheritance affects the body’s response to drugs, a field known as pharmacogenomics (9). By better analyzing a patient’s individual genetic makeup, doctors can prescribe the best drug therapy and the appropriate dosage for the treatment and avoid any unintended side effects (9). Current methods of pharmacogenetics include genotyping for genes involved in the action and metabolism of drugs.
Figure 3: Personalized medicine holds a tremendous amount of potential on targeting difficult diseases.
Furthermore, pharmacogenomics may have important implications in health policy (Fig. 3). Each year in the United States, about 106,000 deaths and 2.2 million serious incidents are caused by adverse drug reactions (ADR) (10). ADRs can also lead to the withdrawal of drugs from the market, leading to millions of dollars in wasted development, trial, marketing, and litigation costs for drug companies and federal grant-giving agencies.
Widespread adoption of personalized medicine may enable more efficient medical trials. Scientists anticipate that by linking genetics and proteomics with the body’s reaction to external chemical agents, researchers could create more powerful medicines and safer drugs, as well as discover appropriate drug dosages, advanced screening for disease, and better vaccines.
Policy and Delivery
Health Policy: Regulating the interface between NIH, academia, and private biotech companies
The actualization of personalized medicine will require effective cooperation between academia and industry through “sharing of data, expertise, resources, and tools” (11). In an example of modern-day collaborative drug development, the NIH Therapeutics for Rare and Neglected Diseases Program (TRND) tests promising compounds by bearing the financial and research burden of the high-risk preclinical development phase, which pharmaceutical companies may be reluctant to undertake (12). Scientists hope that this type of active collaboration will accelerate the development of new therapies for rare diseases and identify molecularly distinct variations of common diseases, leading to new treatments options (12).
Yet within the intersection of federal research agencies, academia, and biotechnology—where individual players have different motivations—conflicts of interest are inevitable. In 1980, the passage of the U.S. “Bayh-Dohl University and Small Business Patent Act” enabled universities, private businesses, and non-profit organizations to retain ownership of an invention from research funded by a federal agency. While the legislation enabled more biotech companies to attract investment capital in science and accelerated commercialization of federally funded inventions, improper proprietary patents may stifle progression of research.
For instance, imagine a tangible intellectual property (IP)—such as a cell line—discovered by a biotech company with federal research funding. The IP may be a lucrative product that the biotech company can patent under Bayh-Dohl Act to develop commercial products; on the other hand, the patent may prohibit laboratories from gaining access to an essential research tool, presenting a potential conflict of interest between proprietary rights and the freedom of research and innovation.
The viability of personalized medicine will require research focused on associations between drug response, genetic variation and drug development (11). This type of research and development will be most effective through cross-institutional collaboration and access to research tools for continuation of research (11). As such, the federal government will have to balance the freedom of research with the protection of proprietary rights.
Conclusion
Although the promise of personalized medicine is exciting, it currently exists only in disconnected components. The advent of the Human Genome Project fueled hopeful speculations, leading to genome-wide studies to connect genetic variations with physical traits. Enterprising new startups have embraced the spirit of personalization through the development of personalized diagnosis, which may become more commonly utilized as a medical diagnostics tool with their acceptance by the FDA. However, the realization of personalized medicine is still hampered by limitations in funding and logistics. In order to bring personalized medicine into reality, scientific development, medical translation, health delivery and health policy must come together in effective forms.
Contact Yoo Jung Kim at Yoo.Jung.Kim.14@dartmouth.edu
References
1. P. M. Visscher, M. A. Brown, M. I. McCarthy, J. Yang, Am J Hum Genet. 90, 7–24, (2012).
2. In Treatment for Leukemia, Glimpses of the Future (2012). Available at http://www.nytimes.com/2012/07/08/health/in-gene-sequencing-treatment-for-leukemia-glimpses-of-the-future.html?pagewanted=all (July 2012)
3. How it works (2012). Available at https://www.23andme.com/howitworks2/ (July 2012)
4. 23andMe personalized DNA test seeks FDA approval (2012). Available at http://www.cbsnews.com/8301-504763_162-57483267-10391704/23andme-personalized-dna-test-seeks-fda-approval/ (July 2012)
5. NINDS Tay-Sachs Disease Information Page (2011). Available at http://www.ninds.nih.gov/disorders/taysachs/taysachs.htm (July 2012)
6. H. Ostrer, Legacy: A Genetic History of the Jewish People (Oxford University Press, New York, ed. 1, 2012). [first edition]
7. A. L. Taylor et al., N Engl J Med. 351, 2049-2057, (2004).
8. J. Kahn, Am J Bioeth. 5, W1-5, (2006).
9. Pharmacogenomics (2011). Available at http://www.ornl.gov/sci/techresources/Human_Genome/medicine/pharma.shtml (July 2012)
10. J. Lazarou, B. Pomeranz, P. N. Corey, JAMA, 279, 1200-1205, (1998).
11. R. Gupta, J. P. Kim, J. Spiegel, and S. M. Ferguson, Personalized Med. 1, 115-124, (2004).
12. M. A. Hamburg F. S. Collins, N Engl J Med, 363, 301-304, (2010).