Soyeon (Sophie) Cho ’24, Biological Sciences, Winter 2021
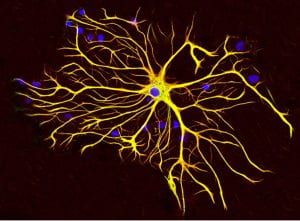
Cover Image: In this tissue culture, an astrocyte cell is surrounded by other cells. Antibodies to the two main proteins, GFAP and vimentin, stain the cells with red and green fluorescent dye, respectively. Since both proteins are widely distributed throughout the astrocyte, the cell appears to be generally yellow. The blue circles represent the nuclei of the cells which contains the cell’s DNA. (Source: Wikimedia Commons)
It is well known that that humans are closely related to the great apes like chimpanzees and gorillas. Specifically, humans and chimpanzees share 98.8% of their DNA, meaning that they only differ by 35 million nucleotides (Varki & Altheide, 2005). However, these remaining 35 million nucleotides do not necessarily determine all the differences between humans and chimpanzees. Many studies have suggested that evolutionary changes in the protein sequence cannot explain all of the differences between humans and chimpanzees, especially in the brain.
A common hypothesis suggests regulatory adaptations through adaptive evolution, which is the evolutionary process that pushes organisms to respond to a changing environment by becoming more “fit.” Regulatory adaptations start as genetic mutations in the binding sites, such that transcription is promoted or hindered by their corresponding regulatory proteins. These mutations change the protein’s binding affinity. If this binding affinity change increases evolutionary “fitness,” the mutations cause adaptive evolution as regulatory adaptations in the DNA. Two selective mechanisms are responsible for adaptive evolution: negative (purifying) selection, which encourages conservation of current traits, and positive selection, which promotes the gain of new characteristics (Vallender & Lahn, 2004). Thus, if increasing the binding affinity of a “beneficial” DNA sequence or decreasing the binding affinity of a “harmful” DNA sequence increases the likelihood of survival and reproduction, regulatory adaptations lead to adaptive evolution over time.
Despite this background, previous studies had not found direct evidence to support this hypothesis by identifying specific regulatory changes in the brain. In a novel study, Liu and Robinson-Rechavi (2020) measured changes in protein binding affinity to provide this missing evidence between brain-related functions and adaptive evolution.
Liu and Robinson-Rechavi utilized a machine learning model and experimental data on transcription factor binding sites (TFBSs). This study differentiates itself from previous studies because it reduces the impact of neutral, nonadaptive mechanisms on positive binding sites (PBSs) for transcription factors (Liu & Robinson-Rechavi, 2020). Gkm-SVM is a support vector machine (SVM), a machine learning model that calculates a linear divider between two groups of data points. It predicts the binding affinity of regulatory DNA elements like transcription factors (Lee, 2016).
First, the researchers used a positive and negative set to train the gkm-SVM. The positive set of gkm-SVM is comprised of the sequences for transcription factor binding sites (TFBS) (Ghandi et al., 2014). These TFBS sequences are derived by chromatin immunoprecipitation sequencing (ChIP-seq), a technique that uses a protein-specific antibody to identify the protein’s binding sites on DNA (Das et al., 2004). The negative set consists of negative-control sequences with the same length. These sets train the gkm-SVM to predict how each “10-mer,” a random 10-nucleotide long sequence, affects the binding affinity of the transcription factor. These predictions are called SVM weights, and a positive or negative SVM weight indicates positive or negative regulatory activity, respectively (Lee et al., 2015).
Secondly, researchers determined whether a TFBS underwent positive selection for different species. Liu and Robinson-Rechavi identified TFBS sequences for the ancestral and focal species using a phylogenetic tree. These sequences were repeatedly entered into the gkm-SVM, producing bell-shaped distributions for the deltaSVM values, or changes in binding affinity from the ancestral to focal sequences. They reasoned that because changes in gene expression are positively selected if they directly increase evolutionary fitness, large absolute values of deltaSVM would indicate that a binding site was positively selected over time (Liu & Robinson-Rechavi, 2020).
Next, the team calculated deltaSVM p-values, the probability that the observed deltaSVMs, or binding affinity changes, were random. According to Liu and Robinson-Rechavi (2020), a binding site with a deltaSVM p-value less than 0.01 was defined as a binding site that underwent positive selection, since the large absolute value of deltaSVM around the optimum would be significant. Therefore, the gkm-SVM would be able to deduce whether an ancestral TFBS underwent positive selection, causing a new optimum of binding affinity for the corresponding focal TFBS.
To validate this method, they tested for positive selection in liver specific TFBSs for a mouse species (M. musculus domesticus) and humans (Homo sapiens). Then, researchers determined the ancestral sequence by comparing equivalent TFBSs for chimpanzees (a sister group) and gorillas (an outgroup). According to both validation studies, the TFBSs with higher binding affinity also shared the characteristics for positively selected binding sites: there was “higher substitution-to-polymorphism ratio in sequence, and lower variance in expression of neighboring genes” (Liu & Robinson-Rechavi, 2020).
After confirming that the gvm-SVM accurately predicts positive selection, they tested binding sites for the transcription factor CTCF, since it exists in 29 cell types for Homo sapiens and 11 tissue types for the M musculus. Using the deltaSVM values for each TFBS, the study indicated that for humans, brain-related cell types are more likely to demonstrate positive selection than others. For example, the highest proportion of positively selected binding sites were found in the choroid plexus epithelial cells and the brain microvascular endothelial cells (Liu & Robinson-Rechavi, 2020). However, the mice showed similar levels of positive selection and adaptive evolution throughout all cell-type CTCF binding sites, including brain-related ones. This observation indicates that high positive selection in regulatory elements in the brain is not a general mammalian trait, but a human-specific characteristic.
Ultimately, this study illustrates that more research is needed into determining the binding site differences between humans and mice, or even humans and other mammals. For example, ChIP-seq data was extracted from cell types for humans and tissues for mice, and some of the cell or tissue types lacked equivalents in the other species (Liu & Robinson-Rechavi, 2020). Nevertheless, it has various implications in the cognitive abilities of humans. It provides strong evidence for high levels of positive selection in the human brain, indicating that gene expression in the brain may be producing characteristics unique to humans (Liu & Robinson-Rechavi). Additionally, some of the brain regions studied here are closely related to cognitive functions, including abnormal signaling and Alzheimer’s. Therefore, it is anticipated that this study will guide future research on not only the evolution of specific genomic regions but also their applications in medicine.
References
Das, P. M., Ramachandran, K., vanWert, J., & Singal, R. (2004). Chromatin immunoprecipitation assay. BioTechniques, 37(6), 961–969. https://doi.org/10.2144/04376rv01
GerryShaw. (2013, November 2). Astrocyte [Photograph]. Wikimedia Commons. https://commons.wikimedia.org/wiki/File:Astrocyte5.jpg
Ghandi, M., Lee, D., Mohammad-Noori, M., & Beer, M. A. (2014). Enhanced regulatory sequence prediction using gapped k-mer features. PLoS computational biology, 10(7), e1003711. https://doi.org/10.1371/journal.pcbi.1003711
Kastritis, P. L., & Bonvin, A. M. J. J. (2013). On the binding affinity of macromolecular interactions: daring to ask why proteins interact. Journal of The Royal Society Interface, 10(79), 20120835. https://doi.org/10.1098/rsif.2012.0835
Lee, D., Gorkin, D. U., Baker, M., Strober, B. J., Asoni, A. L., McCallion, A. S., & Beer, M. A. (2015). A method to predict the impact of regulatory variants from DNA sequence. Nature Genetics, 47(8), 955–961. https://doi.org/10.1038/ng.3331
Lee D. (2016). LS-GKM: a new gkm-SVM for large-scale datasets. Bioinformatics, 32(14), 2196–2198. https://doi.org/10.1093/bioinformatics/btw142
Liu, J., & Robinson-Rechavi, M. (2020). Robust inference of positive selection on regulatory sequences in the human brain. Science Advances, 6(48), eabc9863. https://doi.org/10.1126/sciadv.abc9863
Swiss Institute of Bioinformatics. (2020, December 16). The DNA regions in our brain that contribute to make us human. ScienceDaily. https://www.sciencedaily.com/releases/2020/12/201216085039.htm
Vallender, E. J., & Lahn, B. T. (2004). Positive selection on the human genome. Human Molecular Genetics, 13(suppl_2), R245–R254. https://doi.org/10.1093/hmg/ddh253
Varki, A., & Altheide, T. K. (2005). Comparing the human and chimpanzee genomes: searching for needles in a haystack. Genome research, 15(12), 1746–1758. https://doi.org/10.1101/gr.3737405