Lord Charité Igirimbabazi, Biological Sciences, Spring 2021
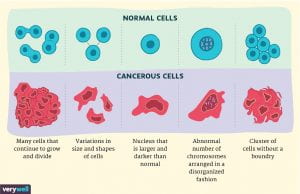
Figure: Shows structural differences between cancer cells and normal cells. Researchers have shown that tumor cells are a mixture of cancer cells and normal cells. Distinguishing malignant cells and nonmalignant cells in the tumor microenvironment is critical for understanding gene expression programs of tumor cells. (Source: Verywell Website)
Single Cell RNA sequencing (scRNA-Seq), a transcriptomic analysis tool, has become important in cancer research since it provides deeper insights to the genetic material or transcriptional profiling of different single cells from a same tissue sample. This sequencing method is significantly different from a widely used sequencing method—bulk RNA sequencing—that gives an averaged gene expression of each gene across all sampling cells (Shalek et al., 2014). Despite major breakthroughs in genome sequencing methods, cancer researchers have struggled to analyze the complex and large datasets emerging from scRNA-Seq of tumor samples in such a way as to distinguish cancer cells from nonmalignant cells since the two cell types coexist in the tumor microenvironment. To address this problem, researcher Ruli Gao and her colleagues from the University of Texas MD Anderson Cancer Center developed a specialized computational tool—CopyKAT (Copy number Karyotyping of Aneuploidy Tumors)—to find potential genetic fingerprints to differentiate cancer cells from normal cells in the tumor microenvironment.
Drawing on the fact that copy number profiles (i.e., a way to represent the number of copies of a particular gene in the genome) of normal cells are often diploid while those of cancer cells are often indicative of aneuploidy which often results in uncontrollable cellular division (Figure1), CopyKAT can infer the genetic makeup of each individual cell in the tumor mass, identifying at a much higher resolution cells with an abnormal number of chromosomes (University of Texas, 2021). This is especially important since the aneuploid cells are the major characteristic of most human tumors. CopyKAT uses integrated Bayesian methods to identify genome-wide aneuploidy events with a resolution of 5Mb from scRNA-Seq data. To achieve this feat, Gao and her colleagues applied CopyKAT to infer the copy number profile of nonmalignant or diploid cells by compartmentalizing individual cells into a number of hierarchical clusters (Gao et al., 2021).
Gao and her colleagues utilized a Gaussian Mixture Model to evaluate the variance of each cluster, and the cluster with a few variances is selected as that containing confident diploid cells to infer the ground-state copy number profiles. However, errors may occur when the tumor sample contains a small number of normal cells or cancer cells possess diploid-like genomes with a few copy number alterations, i.e., genomic alterations to chromosome structure that result in deletions or amplifications of DNA material that often contribute significantly to the progression of cancerous tumors (Gao et al., 2021). As an alternative approach, the team evaluated the diploid status of each single cell one at a time, and a single cell with a higher diploid status was selected as a normal cell to estimate the baseline copy number values.
This tool was applied on 21 different tumors, including pancreatic cancer, triple-negative breast cancer and anaplastic thyroid cancer. CopyKAT separated cancer cells from normal cells successfully with an average of 98% accuracy for all the different datasets (Gao et al. 2021).
Another appealing feature of this computational tool is its ability to analyze the genetic diversity or heterogeneity that arises from cancer cells. Gene expression of cancer cells may differ due to cancer cells’ proliferation or external pressures. Thus, cancer cells can be organized into clonal subpopulations based on the copy number differences, which may give insights about gene expression differences among the cancer subpopulations (Gao et al. 2021). Gao and her colleagues selected and identified the clonal subpopulations by clustering single cell copy number datasets through comparing the copy number profiles of aneuploid tumor cells and diploid cells.
CopyKAT is a promising computational tool in cancer research and clinical oncology. This groundbreaking tool will greatly improve understanding of gene expression differences among tumor cells and how chromosome alterations lead to different cancer phenotypes. With information about the genetic makeup of cancer subpopulations, a number of important clinical questions emerges, such as how cancerous tumors proliferate and evolve, which pathways might or should be targeted, and how successful targeted therapy will be for a particular patient.
References
Gao, R., Bai, S., Henderson, Y.C. et al. Delineating copy number and clonal substructure in human tumors from single-cell transcriptomes. Nature Biotechnology (2021). https://doi.org/10.1038/s41587-020-00795-2
Shalek, A., Satija, R., Shuga, J. et al. Single-cell RNA-seq reveals dynamic paracrine control of cellular variation. Nature 510, 363–369 (2014). https://doi.org/10.1038/nature13437
University of Texas M. D. Anderson Cancer Center. (2021, January 18). New computational tool reliably differentiates between cancer and normal cells from single-cell RNA-sequencing data: CopyKAT enables researchers to gain new insights when analyzing solid tumor samples. ScienceDaily. Retrieved April 12, 2021 from www.sciencedaily.com/releases/2021/01/210118113040.htm