Frankie Carr, Biological Sciences, Spring 2021
Figure 1: A schematic of the double helix of DNA (Source: Wikimedia Commons).
DNA is made of nucleotides, which consist of a deoxyribose sugar bound to a phosphate group along with a nitrogen base, joined together with bonds between their sugars and phosphate groups. Most organisms contain 4 different nitrogenous bases in their DNA: Adenine (A), Thymine (T), Guanine (G), and Cytosine (C). Two different strands of DNA combine to form a double helix structure; specifically, the two strands are joined via hydrogen-bonds between their nitrogenous bases. This “base-pairing” occurs between one purine (adenine or guanine) and one pyrimidine (cytosine or thymine), with A-T pairs (with 2 hydrogen bonds) and C-G (with 3 hydrogen bonds) pairs. Some viruses have been detected to replace their Adenine bases with Diaminopurine (Z) bases, forming Z-T pairs, which have a third stability-providing hydrogen bond (see Figure 2 below).
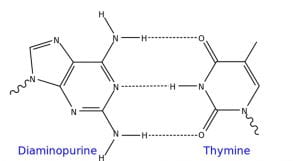
Figure 2: A diagram of the base-pairing that occurs between Diaminopurine (Z) and Thymine (T), hydrogen bonds are indicated by dashed lines (Source: Wikimedia Commons).
The use of Diaminopurine in the cyanobacteria-infecting S-2L Cyanophage was first reported in 1977. Researchers noted that the Z base was capable of 3 hydrogen bonds, which gives the viral DNA a higher melting temperature, thus indicating greater stability than regular DNA (Kirnos et al., 1977). Later, Bailly and Waring showed that DNA containing Diaminopurine instead of Adenine had unique structural features when compared to regular DNA. These features included a more hydrophilic minor groove (due to the additional amino group) and greater stability of DNA in the Z conformer (DNA can exist in one of three conformations, A, B, and Z, with B being the most common) (Bailly & Waring, 1998). Additionally, it was shown that these conformational changes, along with the presence of the second amino group, render the viral DNA unrecognizable and uncleavable by most restriction enzymes, which bacteria often use to recognize and destroy viral DNA. Thus, the switch to Diaminopurine may provide the viruses with increased ability to infect their target bacteria (Szekeres & Matveyev, 1987).
Recently, three studies published in Nature from a team of researchers have shown the biochemical synthesis of Diaminopurine, the method of incorporation of the base into DNA, and the widespread use of these enzymatic pathways in various viruses.
Adenine and Diaminopurine are very structurally similar. The only difference between the two is the additional amino group on the C2 carbon in diaminopurine(see Figure 3).
Figure 3: a comparison between the structures of Adenine (left) and Diaminopurine (right), note the second amino (NH2) group on the right ring of diaminopurine (source: Wikimedia Commons).
The biochemical synthesis of the new nitrogenous base is not simply done by adding the amino group to the carbon. Instead, it was recently shown by Sleiman’s team that the synthesis occurs via a parallel pathway to that of the Adenine synthesis using the enzyme PurZ (named because it is involved in the synthesis of the Purine Z). Adenine biosynthesis occurs via the PurA enzyme, and both enzymes’ products are then shuttled further down the purine synthetic pathway by PurB. This pathway results in the free nucleotide (dAMP or dZMP) that can then be inserted into the DNA during replication (Sleiman et al., 2021).
In the past, researchers have confirmed that viral DNA polymerases can recognize and use Z, but they do not differentiate between using it and A. Since the DNA found in these viruses contains entirely Diaminopurine and no Adenine, there must be some polymerase that preferentially selects Z over A. In exploring this idea, a team of researchers working with those who discovered PurZ found such a polymerase called DpoZ (named because it is a DNA Polymerase that has selectivity for Z) (Pezo et al., 2021).
Two papers from this same team detailed the evolutionary route of the DpoZ and PurZ; the researchers demonstrated that both enzymes are found in many different kinds of bacteriophages, especially those in the Podoviridae and Siphoviridae families. Additionally, they showed that the two enzymes evolved in tandem and possibly originated around 3.5 billion years ago. The researchers theorized that the enzymes likely evolved after some duplication of their adenine-favoring counterparts due to the homology between the two sets of genes (Pezo et al., 2021; Zhou et al., 2021).
The Z nucleotide is not the first modified base to be found in DNA; many organisms, including humans, methylate cytosine bases to form 5-Methylcytosine, which is used to regulate genes in a process referred to as epigenetics (Bailly & Waring, 1998). Even in viral DNA, various methylated bases provide a virus “memory” of its recent hosts. Additionally, various other groups can and can be added onto the four basic nucleotides to serve a variety of purposes (Weigele & Raleigh, 2016). However, Diaminopurine is the first modified base that has been detected that (1) alters the amount of hydrogen bonds in the base pair and (2) originates not from simple modification of a pre-existing base but from its own biosynthetic pathway (Pezo et al., 2021; Sleiman et al., 2021; Zhou et al., 2021).
Future research should be done on the existence of other PurZ and DpoZ homologs in viruses and other organisms to see if another unique base exists and is incorporated into their DNA. Additionally, research should explore the transcription and translation of the viral DNA into mRNA and protein within a host cell. It is likely that the increased hydrogen bonds between Diaminopurine and Thymine will also be present in the interaction between Diaminopurine and Uracil (the Thymine analog seen in RNA and some viral genomes), possibly leading to a slowed or altered transcriptional process.
References
Bailly, C., & Waring, M. J. (1998). The use of diaminopurine to investigate structural properties of nucleic acids and molecular recognition between ligands and DNA. Nucleic Acids Research, 26(19), 4309–4314. https://doi.org/10.1093/nar/26.19.4309
Kirnos, M. D., Khudyakov, I. Y., Alexandrushkina, N. I., & Vanyushin, B. F. (1977). 2-Aminoadenine is an adenine substituting for a base in S-2L cyanophage DNA. Nature, 270(5635), 369–370. https://doi.org/10.1038/270369a0
Pezo, V., Jaziri, F., Bourguignon, P.-Y., Louis, D., Jacobs-Sera, D., Rozenski, J., Pochet, S., Herdewijn, P., Hatfull, G. F., Kaminski, P.-A., & Marliere, P. (2021). Noncanonical DNA polymerization by aminoadenine-based siphoviruses. Science, 372(6541), 520–524. https://doi.org/10.1126/science.abe6542
Sleiman, D., Garcia, P. S., Lagune, M., Loc’h, J., Haouz, A., Taib, N., Röthlisberger, P., Gribaldo, S., Marlière, P., & Kaminski, P. A. (2021). A third purine biosynthetic pathway encoded by aminoadenine-based viral DNA genomes. Science, 372(6541), 516–520. https://doi.org/10.1126/science.abe6494
Szekeres, M., & Matveyev, A. V. (1987). Cleavage and sequence recognition of 2,6-diaminopurine-containing DNA by site-specific endonucleases. FEBS Letters, 222(1), 89–94. https://doi.org/10.1016/0014-5793(87)80197-7
Weigele, P., & Raleigh, E. A. (2016, June 20). Biosynthesis and Function of Modified Bases in Bacteria and Their Viruses (world) [Review-article]. American Chemical Society. https://doi.org/10.1021/acs.chemrev.6b00114
Zhou, Y., Xu, X., Wei, Y., Cheng, Y., Guo, Y., Khudyakov, I., Liu, F., He, P., Song, Z., Li, Z., Gao, Y., Ang, E. L., Zhao, H., Zhang, Y., & Zhao, S. (2021). A widespread pathway for substitution of adenine by diaminopurine in phage genomes. Science, 372(6541), 512–516. https://doi.org/10.1126/science.abe4882